Prof. Dr. Robert Tampé
Goethe University Frankfurt, Institute of Biochemistry, Cellular Biochemistry
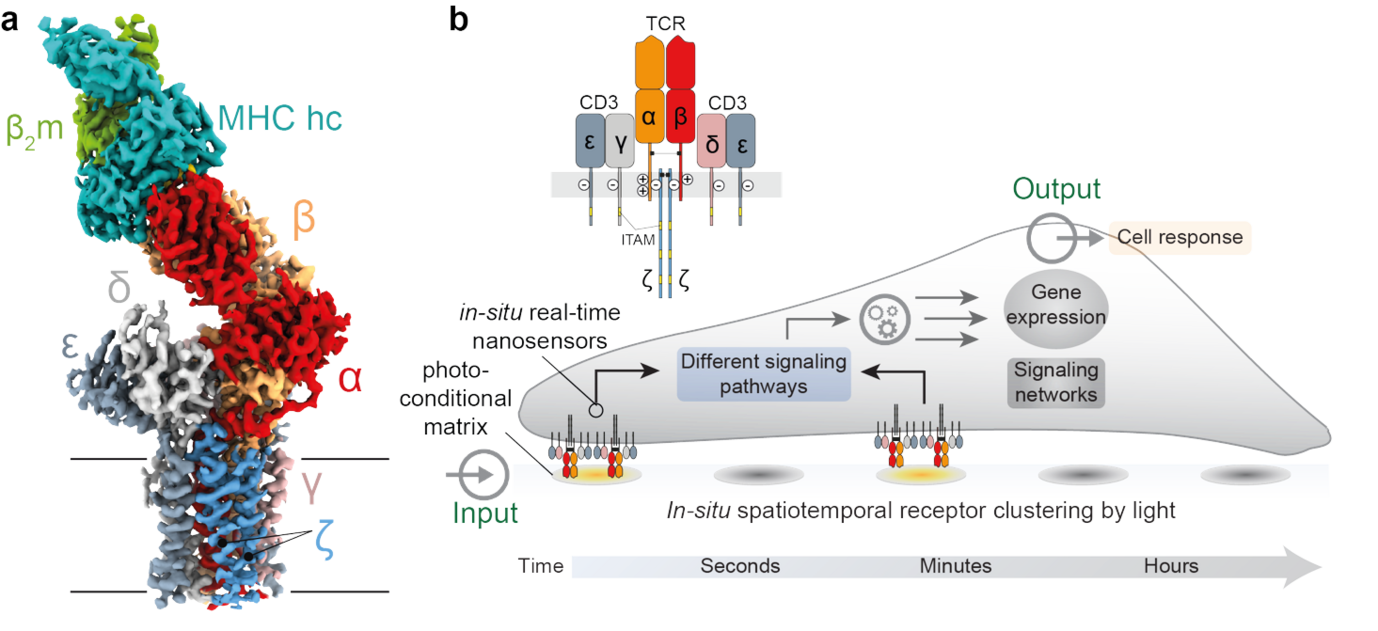
T Cell Receptor Clustering and Early Signaling Triggered by Light
The way receptors, especially T cell receptor (TCR)–CD3 complexes, are organized in space and time plays a fundamental role in triggering immune responses against cancer and infections. While recent advances have been made, the exact details of how receptor organization influences early immune signaling remain unclear. This PhD project aims to address this by developing new light-sensitive tools, called photoactivatable nanotools, which can be controlled using specific wavelengths of light. These tools will allow us to precisely and non-invasively manipulate how receptors cluster and activate, giving us the ability to quickly fine-tune protein networks. We will also create advanced activation systems and analyze them with advanced fluorescence imaging. This project brings together a wide range of disciplines, including molecular biology, biochemistry, biophysics, and cell biology, to explore one of the most important processes in immune response
References:
- Sušac, L., Vuong, M. T., Thomas, C., von Bulow, S., O'Brien-Ball, C., Santos, A. M., Fernandes, R. A., Hummer, G., Tampé, R.* & Davis, S. J.* Structure of a fully assembled tumor-specific T cell receptor ligated by pMHC. Cell 185, 3201-3213 e3219 (2022). https://doi.org:10.1016/j.cell.2022.07.010
- Sanchez, M. F., Faria, S., Frühschulz, S., Werkmann, L., Winter, C., Karimian, T., Lanzerstorfer, P., Plochberger, B., Weghuber, J. & Tampé, R. Engineering mesoscale T cell rdeceptor clustering by plug-and-play nanotools. Adv Mater, e2310407 (2024). https://doi.org:10.1002/adma.202310407
- Sanchez, M. F. & Tampé, R. Ligand-independent receptor clustering modulates transmembrane signaling: a new paradigm. Trends Biochem Sci 48, 156-171 (2023). https://doi.org:10.1016/j.tibs.2022.08.002
- Sanchez, M. F., Dietz, M. S., Muller, U., Weghuber, J., Gatterdam, K., Wieneke, R., Heilemann, M., Lanzerstorfer, P. & Tampé, R. Dynamic in situ confinement triggers ligand-free neuropeptide receptor signaling. Nano Lett 22, 8363-8371 (2022). https://doi.org:10.1021/acs.nanolett.2c03506
- Sanchez, M. F., Els-Heindl, S., Beck-Sickinger, A. G., Wieneke, R. & Tampé, R. Photoinduced receptor confinement drives ligand-independent GPCR signaling. Science 371 (2021). https://doi.org:10.1126/science.abb7657
Contact: tampe@em.uni-frankfurt.de
