Prof. Dr. Michael Kracht, M.D.
Justus Liebig University Giessen, Biomedical Research Centre (BFS)
Rudolf-Buchheim-Institute of Pharmacology
Topics of our group: Chronic inflammatory conditions (such as rheumatoid arthritis, psoriasis or inflammatory bowel disease) affect around 10% of people worldwide. Currently, many new therapies are being introduced, including Janus protein kinase (JAK) inhibitors, monoclonal antibodies directed at cytokines, or cell-based based approaches, but there is still no cure. We follow the idea that recurrent episodes of inflammation are caused by aberrant rewiring of both, signaling and gene expression pathways that together control the formation of specific messenger-ribonucleoprotein particles (mRNPs) encoding inflammatory mediators, rather than by defined genetic mutations. We are therefore pursuing a much more holistic view of the inflammatory gene expression pathway at the molecular level. Our research is focused on identifying the components and regulatory mechanisms that convey signals of gene expression between subcellular compartments. This will enable us to determine the inflammatory proteome in quantitative terms, as well as in terms of time and space..
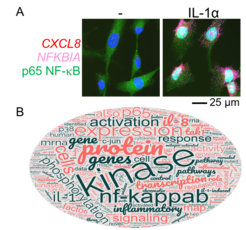
We combine multiple state-of-the-art biochemical, molecular biology, cell biology and bioinformatics methods, including various forms of (epi)genome editing by the CRISPR-Cas9 system, to achieve this. We use human epithelial cells as our major biological models. These cells cover the inner and outer linings of our body and are crucial for our innate immune response. During inflammation, these cells rapidly synthesise and secrete cytokines such as interleukin (IL)-1 and multiple chemokines (Fig. 1A). Together, these factors control the activation and migration of both infiltrating immune cells and resident tissue cells. We challenge the prevailing view and postulate that the cytokine response is primarily controlled by specific processes occurring within the chromatin, rather than by cytosolic signalling pathways. We are therefore analysing the composition of nuclear mRNPs, which are assembled within so-called transcription factories, and the long-range chromatin interactions of these structures. These interactions serve to synchronise the inflammatory gene response in cis (within chromosomes) and in trans (between chromosomes). We also investigate how mRNPs are remodelled when they travel to the cytoplasm and become translated. We believe that gene expression is not simply a linear process but involves multiple levels of feedback control and buffering of mRNA versus protein levels. As examples, we recently demonstrated that targeted mutations of strong inflammatory genomic enhancers not only suppress transcriptional processes but also affect the inflammatory secretome (10).
To gain deeper insights and a more detailed understanding, we employ a comprehensive range of single-cell analyses to examine transcriptional bursting, RNA polymerase II recruitment, and transcriptional elongation. We also investigate post-transcriptional gene regulation, including mRNA decay and protein de novo synthesis. Our interdisciplinary collaborations allow us to apply our expertise in the deep analysis of gene expression at multiple levels, including tissues (Fig. 1B). A recent large-scale study on pathological changes of gene expression in chronic right heart failure is a testament to this approach (1). We also study RNA virus-infected systems, which are extreme examples of cellular stress. In these systems, there is a breakdown in the coordination between nuclear and cytoplasmic gene expression, caused by the phenomenon known as translational shutdown (4, 7, 8).
All of our projects are embedded within a highly active and experienced working group. This environment allows for multiple social and scientific interactions, as well as the rapid acquisition of a broad repertoire of scientific methods and concepts, which are essential for achieving high-ranking first author publications during the PhD or PostDoc work. We are looking for new members for our lab who have successfully finished university degrees in medicine, biochemistry, biology or a related field and who share our specific interest in mechanistically oriented basic medical research, biochemistry, cell biology, molecular imaging, molecular biology and bioinformatics.
We are currently seeking a PhD candidate to join our newly DFG-funded project, entitled "Dissecting functional master-enhancer regimes driving inflammatory stimulation."
For further details see the institute’s webpage and please read the following publications References:
- Jurida L, …, Kracht M. 2024. A common gene signature of the right ventricle in failing rat and human hearts. Nat Cardiovasc Res 3:819-840; (10.1038/s44161-024-00485-1)
- Leib L, …, Kracht M. 2024. The proximity-based protein interaction landscape of the transcription factor p65 NF-kappaB / RELA and its gene-regulatory logics. Biorxiv doi:10.1101/2024.01.03.574021; (10.1101/2024.01.03.574021)
- Priester J, …, Kracht M. 2023. Metabolic labeling and LC-MS/MS-based identification of interleukin-1alpha-induced secreted proteomes from epithelial cells in the presence or absence of serum. STAR Protoc 4:102195; (10.1016/j.xpro.2023.102195)
- Shaban MS, …, Kracht M. 2022. Thapsigargin: key to new host-directed coronavirus antivirals? Trends Pharmacol Sci 43:557-568; (10.1016/j.tips.2022.04.004)
- Mansouri S, .., Kracht M, Savai R. 2022. Cancer genome and tumor microenvironment: Reciprocal crosstalk shapes lung cancer plasticity. Elife 11; (10.7554/eLife.79895)
- Priester J, Dreute J, Kracht M, Schmitz ML. 2022. Post-Transcriptional Control of mRNA Metabolism and Protein Secretion: The Third Level of Regulation within the NF-κB System. Biomedicines 10; (10.3390/biomedicines10092108)
- Shaban MS, …, Kracht M. 2022. Reply to: The stress-inducible ER chaperone GRP78/BiP is upregulated during SARS-CoV-2 infection and acts as a pro-viral protein. Nature Communications 13; (10.1038/s41467-022-34066-2)
- Shaban MS, …, Kracht M. 2021. Multi-level inhibition of coronavirus replication by chemical ER stress. Nat Commun 12:5536; (10.1038/s41467-021-25551-1)
- Meier-Soelch J, Mayr-Buro C, Juli J, Leib L, Linne U, Dreute J, Papantonis A, Schmitz ML, Kracht M. 2021. Monitoring the Levels of Cellular NF-κB Activation States. Cancers 13; (10.3390/cancers13215351)
- Weiterer SS, .., Kracht M. 2020. Distinct IL-1alpha-responsive enhancers promote acute and coordinated changes in chromatin topology in a hierarchical manner. EMBO J 39:e101533; (10.15252/embj.2019101533)
- Kracht M, Muller-Ladner U, Schmitz ML. 2020. Mutual regulation of metabolic processes and proinflammatory NF-kappaB signaling. J Allergy Clin Immunol 146:694-705; (10.1016/j.jaci.2020.07.027)
- Mayr-Buro C, …, Kracht M. 2019. Single-Cell Analysis of Multiple Steps of Dynamic NF-kappaB Regulation in Interleukin-1alpha-Triggered Tumor Cells Using Proximity Ligation Assays. Cancers (Basel) 11; (10.3390/cancers11081199)
- Meier-Soelch J, …, Kracht M. 2018. RNAi-Based Identification of Gene-Specific Nuclear Cofactor Networks Regulating Interleukin-1 Target Genes. Frontiers in Immunology 9; (ARTN 77510.3389/fimmu.2018.00775)
- Poppe M, …, Kracht M. 2017. The NF-kappaB-dependent and -independent transcriptome and chromatin landscapes of human coronavirus 229E-infected cells. PLoS Pathog 13:e1006286; (10.1371/journal.ppat.1006286)
- Schmitz ML, Kracht M. 2016. Cyclin-Dependent Kinases as Coregulators of Inflammatory Gene Expression. Trends Pharmacol Sci 37:101-13; (10.1016/j.tips.2015.10.004)
- Tenekeci U, …, Kracht M. 2016. K63-Ubiquitylation and TRAF6 Pathways Regulate Mammalian P-Body Formation and mRNA Decapping. Mol Cell 62:943-57; (10.1016/j.molcel.2016.05.017)
- Jurida L, …, Kracht M. 2015. The Activation of IL-1-Induced Enhancers Depends on TAK1 Kinase Activity and NF-kappaB p65. Cell Rep 10:726-739; (10.1016/j.celrep.2015.01.001)
- Handschick K, …, Kracht M. 2014. Cyclin-dependent kinase 6 is a chromatin-bound cofactor for NF-kappaB-dependent gene expression. Mol Cell 53:193-208; (10.1016/j.molcel.2013.12.002)
- Ziesche E, …, Kracht M. 2013. The coactivator role of histone deacetylase 3 in IL-1-signaling involves deacetylation of p65 NF-kappaB. Nucleic Acids Res 41:90-109; (10.1093/nar/gks916)
- Rzeczkowski K, …, Kracht M. 2011. c-Jun N-terminal kinase phosphorylates DCP1a to control formation of P bodies. J Cell Biol 194:581-96; (10.1083/jcb.201006089)
- Schmitz ML, …, Kracht M. 2011. Signal integration, crosstalk mechanisms and networks in the function of inflammatory cytokines. Biochim Biophys Acta 1813:2165-75; (10.1016/j.bbamcr.2011.06.019)
- Weber A, Wasiliew P, Kracht M. 2010. Interleukin-1 (IL-1) pathway. Sci Signal 3:cm1; (10.1126/scisignal.3105cm1)
- Gaestel M, Kotlyarov A, Kracht M. 2009. Targeting innate immunity protein kinase signalling in inflammation. Nat Rev Drug Discov 8:480-99; (10.1038/nrd2829)
